About
I am a postdoctoral researcher in Proteomics and Systems Biology at the Institute of Molecular Virology and Cell Biology of the Friedrich-Loeffler-Institut. My research interests focus on bioinformatics, especially omics data and its statistical analysis. My work revolves around analyzing NGS-based genomics and quantitative mass spectrometry-based proteomics data. I consider myself both collaborative and adaptable. I am always eager to tackle new and different projects. I am very enthusiastic about sharing and developing my data visualization skills as a means towards better science communication. I am an open-minded person and enjoy fostering great relationships with my colleagues.
Download my curriculum vitae.
- Bioinformatics
- Data visualization
- Proteomics
- Genomics
- Transcriptomics
- Next generation sequencing
- Network & Systems biology
-
PhD in Bioinformatics & Molecular Biology, 2024
JGU University
-
M.Sc. in Omics Data Analysis, 2017
University of Vic
-
B.Sc. in Biochemistry, 2016
University of Barcelona
Skills
Advanced
Advanced
Good
Advanced
Advanced
Good
Good
Good
Beginner
Experience
In brief:
- Postdoctoral researcher with Dr. Falk Butter
- Genomics (RNAseq, ChIPseq) and proteomics data management, analysis and integration
- Project 1: Developmental proteome quantification in Xenopus species
- Project 2: Leishmania infectome characterization
In brief:
- PhD candidate with Dr. Falk Butter
- Genomics (RNAseq, ChIPseq) and proteomics data management, analysis and integration
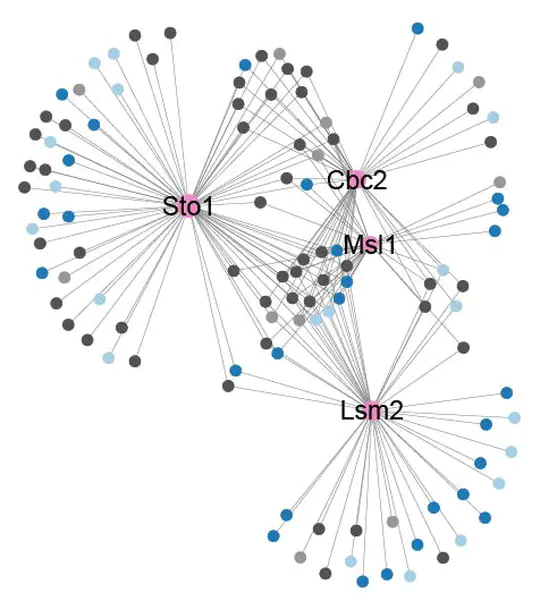
- Project 1: Characterization of the RNA binding proteins interactome in yeast
In brief:
- Research assistant with Dr. Falk Butter
- Genomics (RNAseq, ChIPseq) data management, analysis and integration
In brief:
- MSc student with Dr. Narcs Fernandez-Fuentes
- Development of a scoring function for assessing transcriptome differential expression on Lolium Perenne
In brief:
- B.Sc. student with Dr. Robert Konrat
- Recording and analysis of NMR spectroscopic data
Accomplishments
Projects
Featured Publications

Mass spectrometry-based proteomics data was analyzed to characterize novel interactors for RNA-binding proteins. Network and systems biology strategies revealed concurrent interaction binding patterns and novel functionalities for such proteins.